VPDB TutorialI. Overview: |
VPDB is a viral protein relational database either experimentaly determined X-ray diffraction OR NMR methods. VPDB provides detailed information about viral protein structure II. How to use VPDB Here you can select the "PDBID OR Keywords" Option for database query and then you can inter either PDBID or keywords in the given text box as a input. You can choose Protein Name, Source Of Organism, Virus Name etc. as a keyword search and click into the search button as shown in Fig. 1 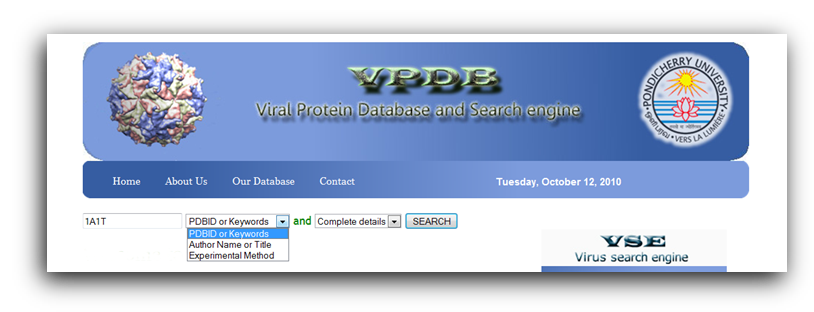 Figure 1. Input page of VPDB as a PDBID OR Keywords Result Page After you press "Submit" on the input page, search tool will search for the HITS that are similar to your input PDBID OR Keywors. The list of hits you can find in the result page. Select the desired one from the list. Use mouse click on PDBID to know the detailed informtion about the selected protein. 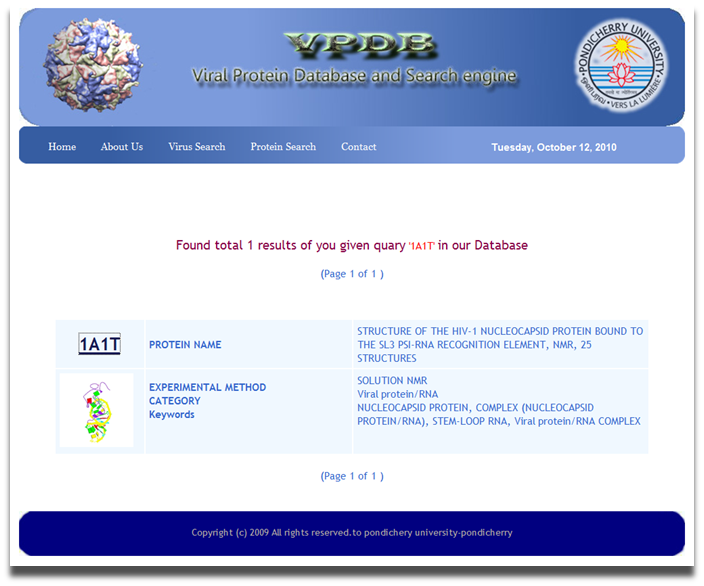 Figure 2. HIT page of PDBID OR Keywords Result Page 0f selected Protein After you Select the desired one from the list. You can find the detailed information in the result page. You can find more informtion on the given Tab options. 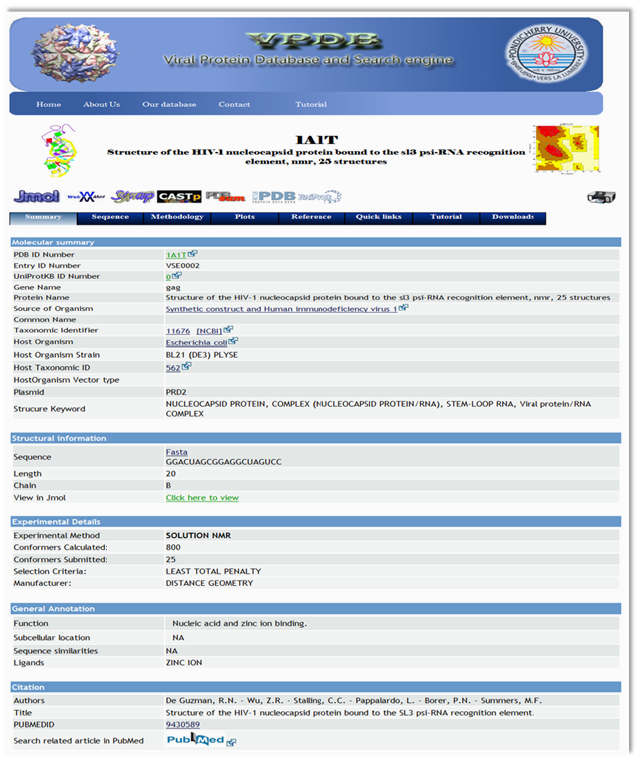 Figure 3. Result page of 1A1T II. Input as a Journal Title OR Author name Here you can select the "Input as a Journal Title OR Author name" Option for database query and then you can inter either Journal Title or Author name in the given text box as a input and click into the search button as shown in Fig. 4 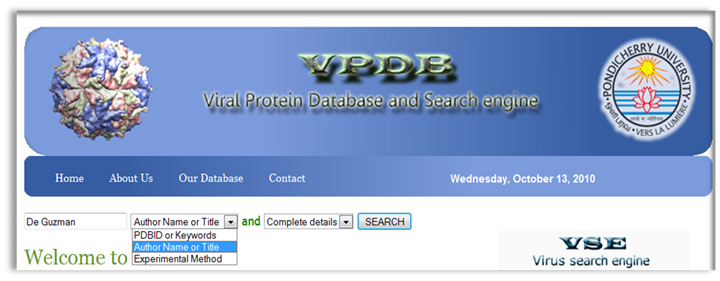 Figure 4. Input page of VPDB as a Journal Title OR Author name Result Page After you press "Submit" on the input page, search tool will search for the HITS that are similar to your input Journal Title OR Author name. The list of hits you can find in the result page. Select the desired one from the list. Use mouse click on PDBID to know the detailed informtion about the selected protein. 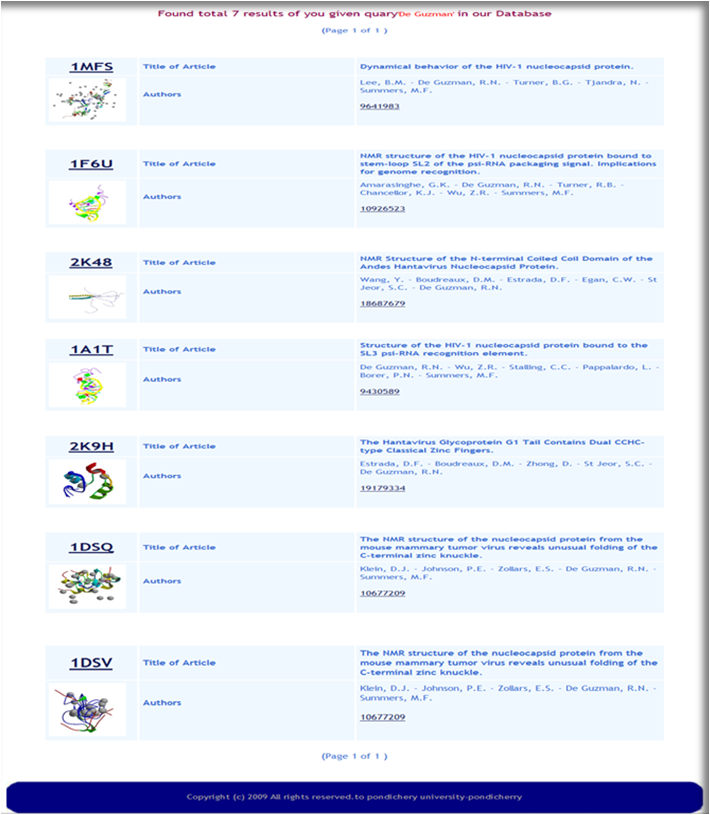 Figure 5. HIT page of Journal Title OR Author name Result Page 0f selected Protein After you Select the desired one from the list. You can find the detailed information in the result page. You can find more informtion on the given Tab options.  Figure 6. Result page of 1A1T III. Input as a Experimental method Here you can select the "Input as a Experimental method" Option for database query and then you can inter the experimetnal method name e.g.<> X-RAY DIFFRACTION, SOLUTION NMR, SOLID-STATE NMR, FIBER DIFFRACTION OR ELECTRON MICROSCOPY in font color="red"> the given text box as a input and click into the search button as shown in Fig. 7  Figure 7. Input page of VPDB as a Experimental method Result Page of Experimental method After you press "Submit" on the input page, search tool will search for the HITS that are similar to your input "Experimental method". The list of hits you can find in the result page. Select the desired one from the list. Use mouse click on PDBID to know the detailed informtion about the selected protein.  Figure 8. HIT page of Experimental method Result Page 0f selected Protein After you Select the desired one from the list. You can find the detailed information in the result page. You can find more informtion on the given Tab options.  Figure 9. Result page of SOLUTION NMR |